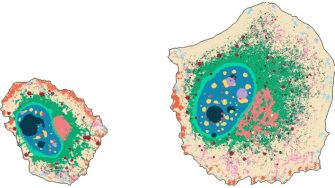
Machine learning to chart intracellular landmarks
Researchers develop an AI tool – with high-resolution images showing many proteins inside human cell lines – to generate cellular maps with intricate detail of the organization of organelles and other cellular landmarks.